Connection to database in IM Browser
IM Browser was designed to visualize interaction data stored in a relational database.
It was tested with MySQL and Oracle databases.
The implementations at DroID and the Finley lab web site connect by default to DroID and C.jejuni databases.
To connect to
a different database select option "Connection" -> "New". IM Browser will prompt
for connection information (see image below). Enter host name (enter
"localhost" or "127.0.0.1" for a MySQL database installed on your
computer), database name, login name and password. Select database type
(MySQL or Oracle).
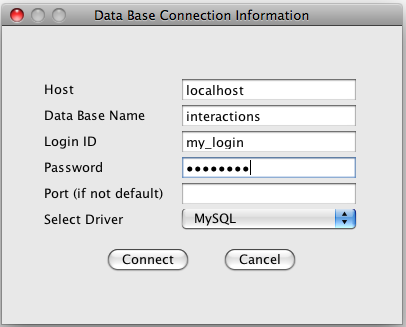
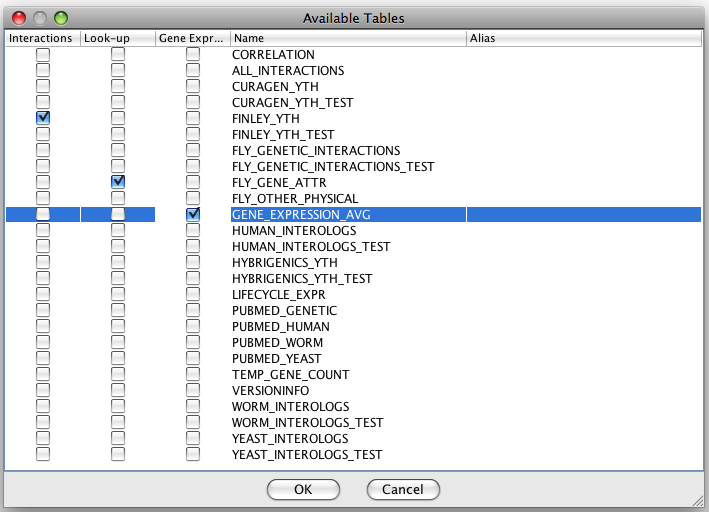
After IM Browser successfully connects to the
database it will display the list of available tables. You have to
specify tables with interactions, and optionally specify a table with
gene attributes (called a lookup table) and a table with gene expression data.
You can have any number of interaction tables. Look-up and expression
tables are optional.
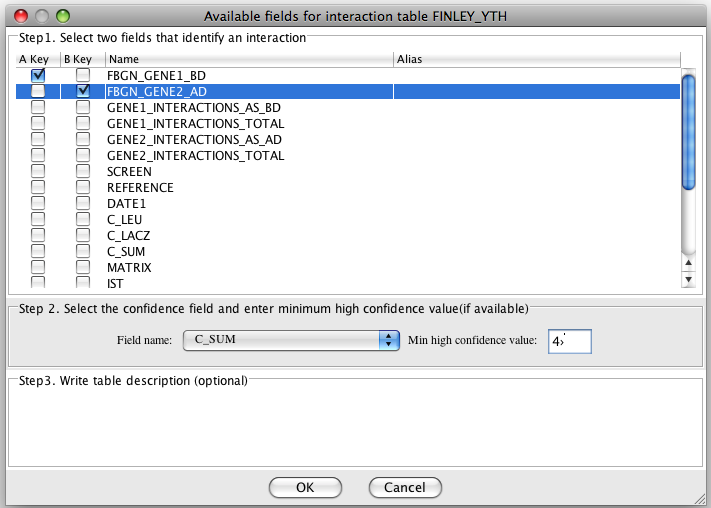
For an interaction table you need to specify which
two fields identify the interaction (they are either the fields with
gene/protein ID, or the fields with gene/protein symbol). Other fields
are considered as interaction attributes. In our database some
interaction tables have dataset-specific confidence score. If your data contains the
confidence score and you want to add/filter the graph by high/low
confidence, you need to specify the field with the confidence score and
enter the minimum value for the high confidence.
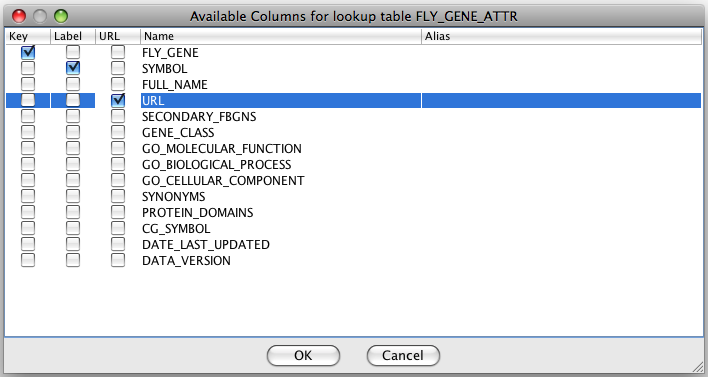
For the lookup table
you need to specify the field with gene/protein IDs. If you want the
node label to be pulled from the lookup table, you need to specify the
field with the node labels.
For the expression table you need to specify
the field with gene/protein IDs.
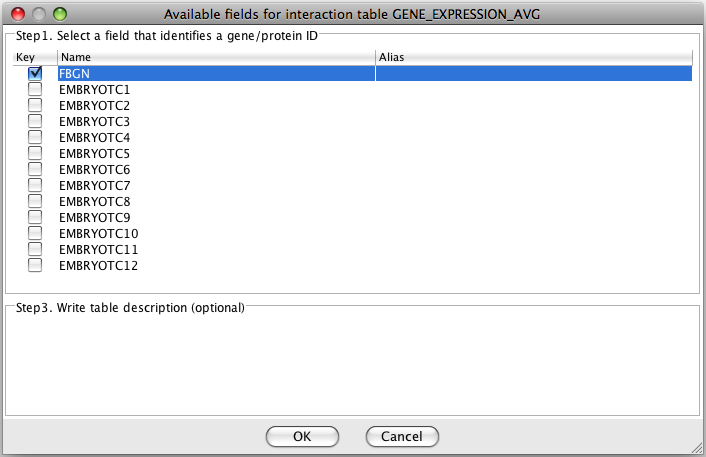
You can save the database configuration
(you will be prompted for it), and load it again through
"Connection"-"Load" the next time you use IM Browser with your
database.
For questions, comments, suggestions feel free to contact Lana
Pacifico (lpacifico at wayne dot edu).





